Why we need more longitudinal aging data
Share on:“We need more human longitudinal data to solve aging” — that’s something you’ve probably heard multiple times before. But why? Wouldn’t cross-sectional data suffice?
It turns out that it’s impossible to answer some of the most intriguing questions about human aging without longitudinal data. Cross-sectional data describe how aging affect an age cohor of people, maybe a few of them in a row. In the best case scenario, we may have a few longitudinal time points for each person, which is, for example, the case for the UK BioBank — of course, you wouldn’t bother a healthy person to get medically re-examined multiple times a week but once a few years it is tolerable. We may sample multiple age cohorts cross-sectionally but they would not enroll a single patient twice. To sum up all that, we may have a few snapshots of how human aging may look like on average, but not dynamically for a given person like you or me.
To illustrate the issue further, we can use a sketch below:
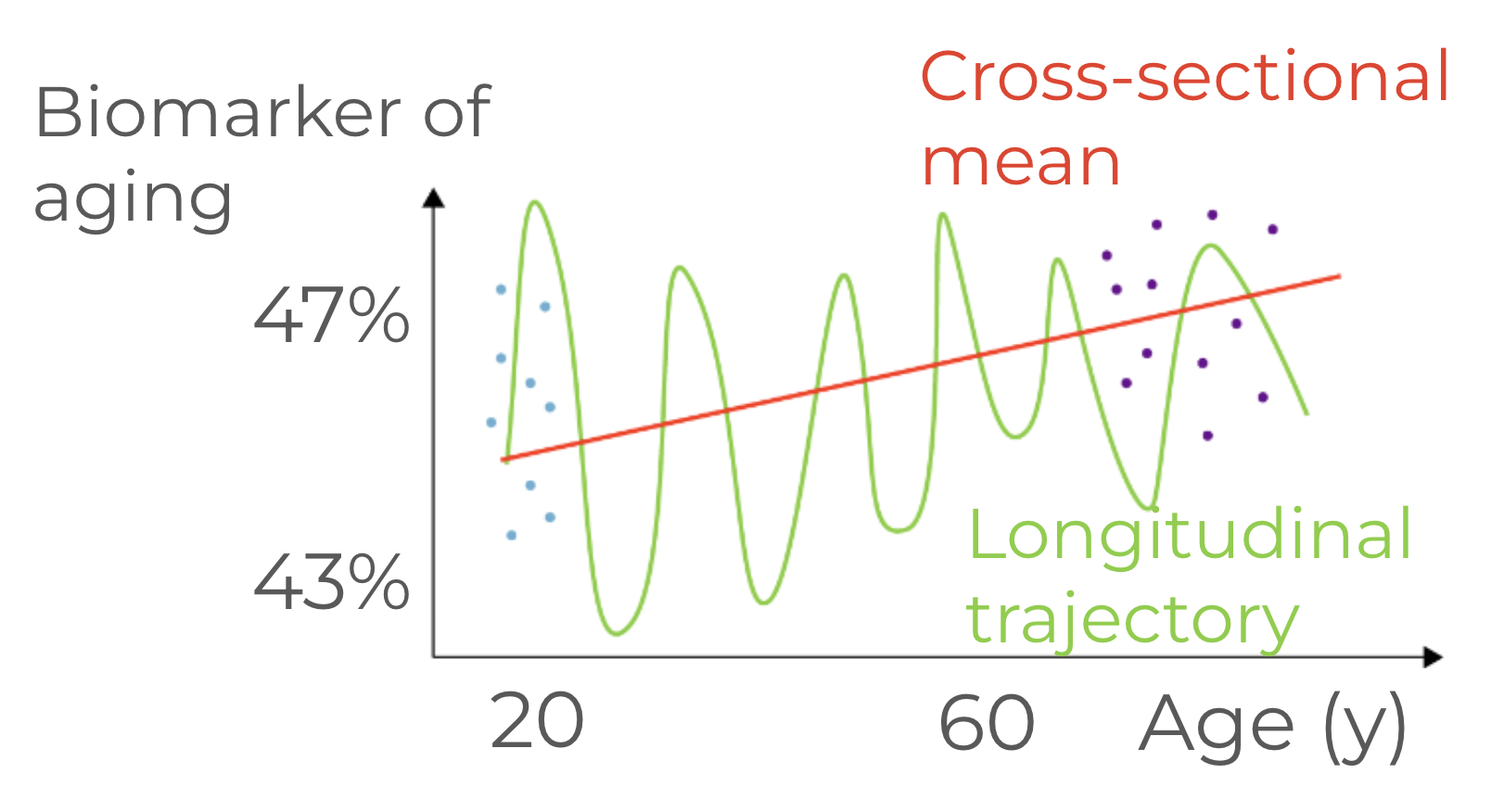
If we simply correlate two cross-sectional snapshots, e.g. young vs old, and identify some biomarker that’s changing between the two in the cross sectional data, we’d end up with some significant correlation between the biomarker and age. The absolute change of the biomarker may not be even prominent — only a few percent changing consistently between young and old would make this biomarker pop up as a predictor of age. The problem arises when we take into account the variability of the biomarker across the cohort, and longitudinally for a single patient — as you see in the sketch, the variability is strong, and there are unlucky young patients who look worse than the most fit old patients. At the same time, even for a single person, we see gigantic fluctuations from time to time which exceed the average change between the young and the old cohorts. The sketch is surprisingly common among the available biological datasets — you can contemplate about complete blood cell counts, biochemical blood tests, e.g. glucose levels, etc. Or, you can consider DNA methylation data and various epigenetic clocks build on that signal — it’s been often noticed that the measurements don’t make much sense for each individual person, and the error from one measurement to another would exceed decades.
For complete blood cell counts, we’ve published two papers (Pyrkov et al. 2021), (Avchaciov et al. 2022) showing that the regimes of longitudinal aging in mice and humans seem to differ qualitatively. For humans, given enough longitudinal data, we could show that the loss of resilience, i.e. the ability to recover to the norm after a stress, is being lost as we age and leads to a prediction that by 120-150 years humans would turn completely unstable, hence that would set the upper bound of human lifespan. Humans turned out to be stable for most of their life until almost the very end, whereas mice seemed to be unstable from the beginning. From the dynamics standpoint the longitudinal data help disentangle the types of aging dynamics — stochastic vs programmatic (many caveats, of course). Assuming a stochastic scenario would imply that the autocorrelation properties of longitudinal signals have a limited memory time (recovery time), and after an organism has recovered from a stress, it shouldn’t affect it too much afterwards. Assuming a programmatic scenario would, on the contrary, imply that there’s something driving those changes in time in a consistent manner, and there may be a scenario where we find some long-term autocorrelations between seemengly independent states & transitions.
It’s nearly impossible to disentangle the two given the longitudinal data we have collected so far for humans. In the future, though, it should become clearer which type of dynamics is realized in practice. Given orthogonal supporting evidence from single-cell variability in DNA methylation and transcriptomes, I’m more personally convinced at this moment that the stochastic changes dominate in most of scenarios (there may be exceptions, of course). Though, we’re far from having enough of high quality longitudinal data to tell for sure which changes are stochastic, and which are programmatic. As always, the reality is most likely somewhere in between — and there’s an intricate interplay of wear and tear, plus genetic predisposition to longevity determining the average longitudinal trajectory. We need more longitudinal aging data!
References
- T.V. Pyrkov, . . ., A.E. Tarkhov, . . ., P.O. Fedichev. Longitudinal analysis of blood markers reveals progressive loss of resilience and predicts ultimate limit of human lifespan. Nat Commun 12, 2561 (2021). 10.1038/s41467-021-23014-1
- K. Avchaciov, . . ., A.E. Tarkhov, . . ., P.O. Fedichev. Unsupervised learning of aging principles from longitudinal data. Nat Commun 13, 6529 (2022). 10.1038/s41467-022-34051-9